The major focus of our laboratory is to elucidate the biological role of lysine methylation in the modulation of intracellular signaling pathways
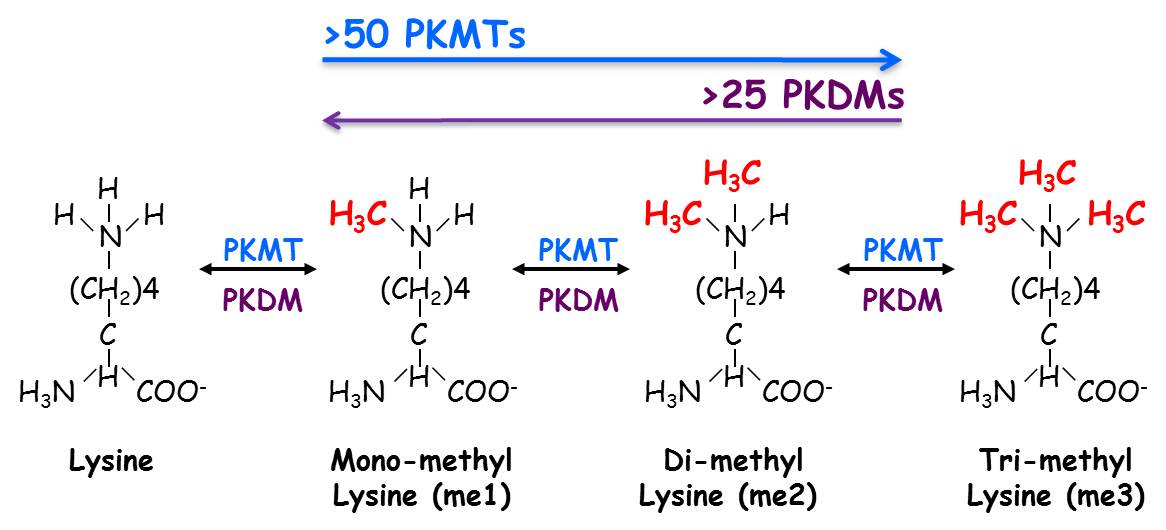
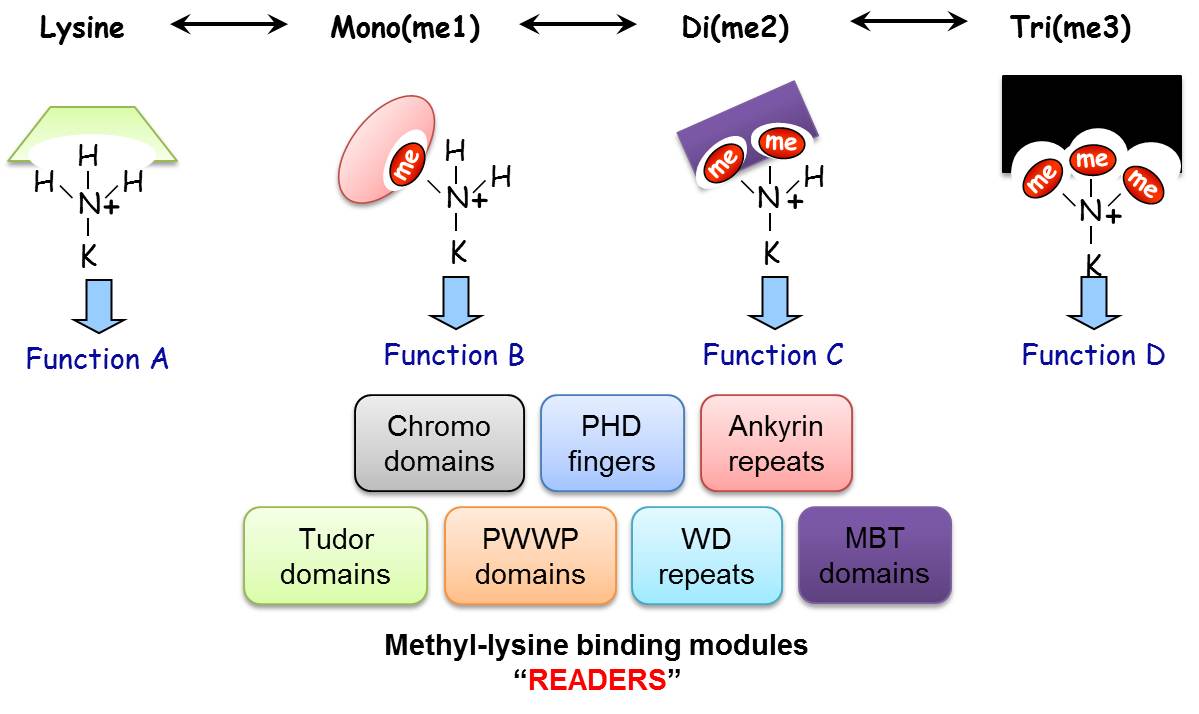
Biological systems respond to extracellular and intracellular signals in myriad ways by activating diverse signaling pathways. Reversible covalent post-translational modifications (PTMs) such as phosphorylation, acetylation and methylation are critical modulators of these signaling pathways. While phosphorylation is the most extensively studied PTM, lysine methylation is emerging as a key player in regulating intracellular signaling pathways. The methylation of lysine residues is performed by protein lysine (K) methyltransferases (PKMTs). It is estimated that in the human proteome there are ~50 PKMT and greater than 25 demethylases (PKDMs) which can reverse the process and remove the methyl mark away. A lysine residue can be mono, di or tri methylated (Left). The degree of methylation is enzyme specific and can define a distinct biological outcome by recruiting specialized regulatory factor, named “readers” that specifically recognize distinct modification in a state (degree of methylation) and in a sequence dependent manner (Right).
 |
 |
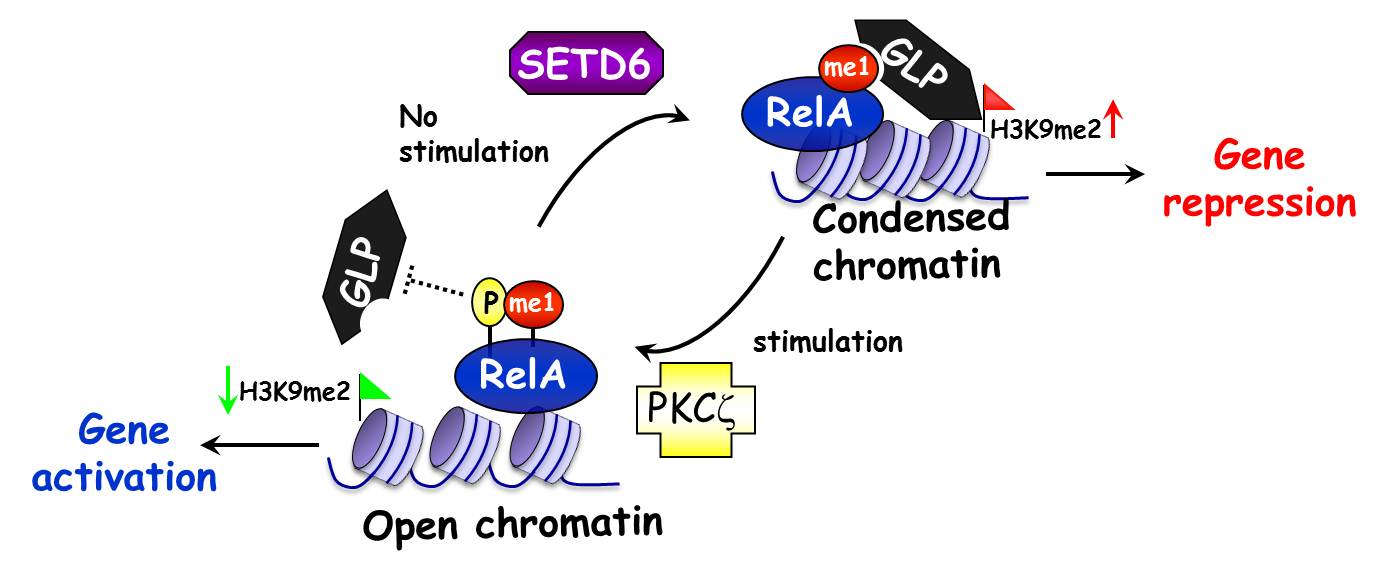
We have recently shown that the transcription factor RelA, a Nuclear Factor-KappaB (NFKB) family member, is methylated by the novel PKMT SETD6. This work described the first known methylation signaling cascade, which is initiated by site-specific mono-methylation of chromatin-associated RelA at the lysine at position 310 (RelAK310me1). We found that SETD6 attenuates RelA transcriptional activity by recruitment of the GLP methyltransferase, which in turn promotes an H3K9me2 silent chromatin state on NFkB occupied genes to locally condense the chromatin and repress transcription of RelA target genes. Depletion of SETD6 and RelAK310me1 levels led to an increase in cell proliferation and a dramatic elevation in secretion of more than 20 inflammatory cytokines both in primary and transformed cells. In response to stimulation with TNFalpha and LPS, phosphorylation of RelA at serine 311 (RelAS311ph) by PKCzeta physically blocks the interaction between GLP and RelAK310me1, leading to transcription activation (Leve et al, Nature Immunology, 2011). This work represents a new paradigm of how integrated crosstalk between modifications on transcription factors and histones governs gene expression programs.
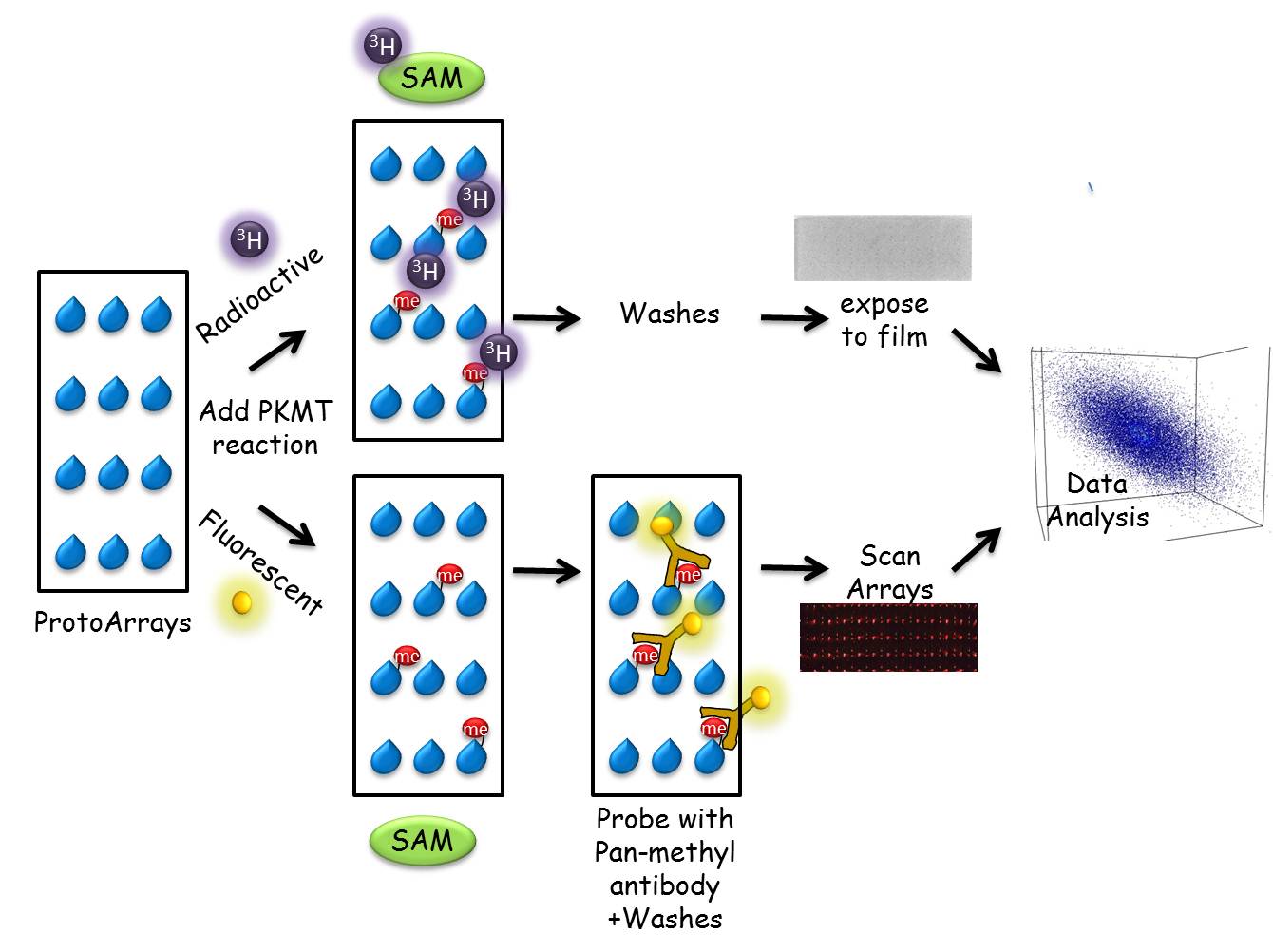
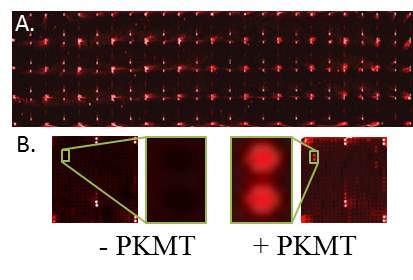
In recent years, lysine methylation has been studied in depth in the context of histones. In recent years there is a growing appreciation that non-histone proteins are also subjected to lysine methylation with a clear role in the regulation of oncogenic and cell differentiation processes. Most studies aiming to identify lysine methylation events have focused on only a few target proteins via a candidate-based approach. We have developed a unique proteomic methodology to identify new substrates for different PKMTs. Using a ProtoArray-based proteomic platform, we demonstrated that more than 9500 unique substrates can be screened in a single experiment in a reproducible and efficient manner (Levy et al, Epigenetics & Chromatin, 2011). Employing this system, we identified 118 new targets for SETD6 and 321 substrates for the related PKMT SETD7. As part of our research program, we will continue to harness and develop protein and peptide array proteomic tools towards the goal of understanding how novel methylation signaling networks impact cancer pathways and cell differentiation programs.
 |
 |
|---|---|
| ProtoArray System: human proteins printed in duplicate on a glass slide allows the screening of more than 9500 potential substrate in a single experiment. | A) full ProtoArray image probed with a pan-methyl antibody after a PKMT reaction. B) Differential methylation signal (green boxes) from arrays incubated without (left) or with a PKMT (right).The red signal represents a positive methylation event. |
Current research directions in the lab are:
- To characterize the biochemical and biological functions of SETD6
- To define the role of lysine methylation in modulating in a variety of biological processes and disease models, with the focus on oncogenic and cell differentiation processes.
- To identify new methylation on histone and non-histone proteins; define the molecular mechanisms by which these methyl marks are generated and transduced; to unravel the biological functions of the methylation events.
- To further develop peptide and protein microarrays technologies focusing on protein lysine methylation.